SDG first steps walkthrough
Workspace
The workspace is the container of a project and contains everything needed work with a genome.
To create a workspace from scratch, first import pysdg.
import pysdg as SDG
Then instanciate a WorkSpace() and save the instance in a variable.
ws = SDG.WorkSpace()
This is the the object where everything will to be stored, the different components of the workspace can be accessed using dot notation.
The main components of the workspace are the graph (ws.sdg), the datastores with their corresponding mappers and the KmerCounters (ws.kci) and most frequently all this data is accessed using NodeViews.
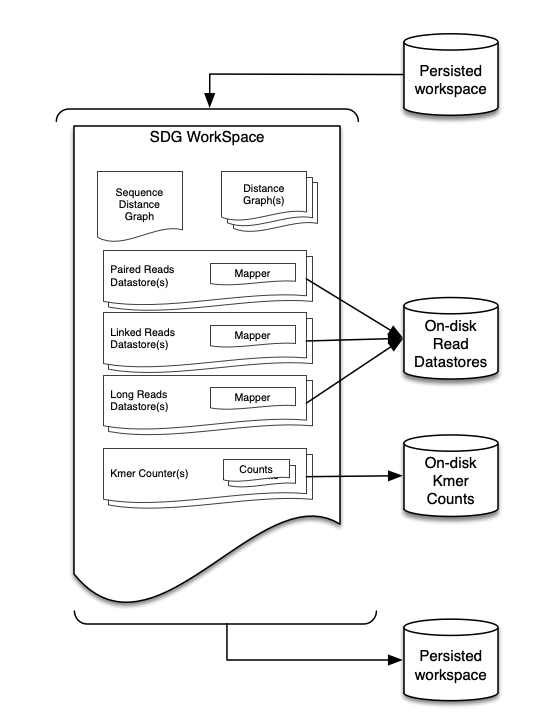
A workspace can be saved to disk and loaded from disk which allows sessions to be preserved. In SDG the processes that are applied to a workspace and it’s components live in memory, so if you want to persist the work that you have done in your graph you’ll need to save a copy of the workspace to disk which can be done using;
>>> ws.dump_to_disk("./persisted_workspace.sdgws")
The session can be restored by loading the workspace instead of rebuilding it from scratch every time;
## Create a new Workspace
>>> restored_ws = sdg.WorkSpace()
>>> restored_ws.load_from_disk("./persisted_workspace.sdgws")
Now restored_ws contains all the same elements as ws.
Please note that some of the components that are derived from the data (like graph kmer indexes) are not persisted in the session and need to be recomputed after loading the workspace from disk.
ws.sdg
SDG is graph based adn each workspace has a main graph where all the data is referenced to. SDG has two types of graphs, sequence digraphs (sdg) and distance graphs (dg). Sequence digraphs are composed of nodes and edges, the nodes store the sequences and the edges represent distances between nodes (distance is the d in sdg). Negative distances indicate overlaps and positive distances indicate N gaps. SDG sequences are stored in their canonical form in the nodes. Distance graphs (dg) only store distances (links) and a reference to the nodes in the main sdg, this type of graph is used to store the results of calculations.
To import a graph to an empty workspace you can load it directly from a gfa file;
>>> ws.sdg.load_from_gfa('./graph.gfa')
Once the graph is loaded, all the information can be accessed directly from the workspace using dot notation or better using Nodeviews() to access the information from the perspective of a node.
Now that the main graph is in the workspace, other data can be ingested by the workspace. SDG supports generic datatypes like paired reads, long reads (Nanopore/PacBio) and linked reads (10x).
Raw data is converted to DataStores so SDG can store an indexed version of the data on disk. This allows the framework to have quick access to the entire dataset without the need to have the entire dataset in memory. This means that workspaces can be bigger than the available RAM. Datastores are type specific, each available data type is stored according to it’s main characteristic.
Datastores can be created using the python API or the command line utilities. For example, to create a paired end datastore using the API.
>>> SDG.PairedReadsDatastore_build_from_fastq("prds.prds", "./pe-reads_R1.fastq", "./pe-reads_R2.fastq", "pe_reads")
the .PairedReadsDatastore_build_from_fastq() method creates a paired read datastore from the fastq files and stores the datastore on the disk ready to be attached to a workspace.
>>> ws.add_paired_reads_datastore("./prds.prds", "PE")
.add_paired_reads_datastore() attaches the datastore to the workspace and names it PE in this case. Attached datastores can be listed using the .list_<datatype>_datastores() method, in this case:
>> ws.list_paired_reads_datastores()
('PE',)
An alternative method of creating datastores is using the command line interface, in this case sdg-datastore
$ sdg-datastore make -t paired -o cli-prds.prds -1 ./pe-reads_R1.fastq -2 ./pe-reads_R2.fastq
This will create a datastore that can be attached in the same way to the workspace;
>>> ws.add_paired_reads_datastore("./cli-prds.prds", "cli-PE")
>>> ws.list_paired_reads_datastores()
('PE', 'cli-PE',)
In a similar way, linked read and long read datastores as well as kmer counters can be attached to the workspace;
## Create linked reads ds and attach it to the ws
>>> SDG.LinkedReadsDatastore_build_from_fastq("./lirds.lirds", "li_reads", "./child/child-link-reads_R1.fastq.gz", "./child/child-link-reads_R2.fastq.gz", SDG.LinkedReadsFormat_raw, readsize=250, chunksize=10000000)
>>> ws.add_linked_reads_datastore("./lirds.lirds", "LI")
>>> ws.list_linked_reads_datastores()
('LI',)
## Create long reads ds and attach it to the ws
>>> SDG.LongReadsDatastore.build_from_fastq("./lords.lords", "longreads_pe", "./child/child-long-reads.fastq")
>>> ws.add_long_reads_datastore('./lords.lords', 'LO')
>>> ws.list_long_reads_datastores()
('LO',)
## Create kmer counter collection in the ws
>>> ws.add_kmer_counter("kmers-PE-k27", 27)
>>> ws.get_kmer_counter("kmers-PE-k27").add_count("PE", ["./pe-reads_R1.fastq", "./pe-reads_R2.fastq"])
>>> ws.list_kmer_counters()
('kmers-PE-k27', )
>>> ws.get_kmer_counter("kmers_noncanonical").list_names()
("PE",)
Now all data is attacched to the workspace and can be usad from within it.
The next thing to do is to indicate SDG how the data in the datastores related to the graph, this is how we want to map the data from the datastores to the nodes.
Each datastore in the workspace contains a mapper object, the mapper is used to map the data to the nodes of the graph.
The mapper deals with mapping the data back to the nodes and the result of mapping the data is a collection of Mappings.
Mappers for each data type have a particular way of mapping the data back to the node and produces different mappings. For example to map long reads from the datastore;
## set the mapper k to 15
ws.get_long_reads_datastore("long_pe").mapper.k=15
## execute the read mapping
ws.get_long_reads_datastore("long_pe").mapper.map_reads()
Alternatively a pointer to the mapper can be constructed to make the code more readable, the mapper is pointed by the variable but is still contained within the datastore in the mapper.
## pointer to the long read mapper
lorm = ws.get_long_reads_datastore("long_pe").mapper
## set k and map the reads
lorm.mapper.k=15
lorm.map_reads()
the resulting mappings are stored inside the mapper in the datastore, for example if you want to check which nodes were mapped to the 1000th read you can user the get_raw-mappings_from_read() function but the best way to access the mappings is using NodeViews (see next section).
>>> for m in lorm.get_raw_mappings_from_read(1000):
>>> .... print (m)
LongReadMapping 1000 (0:8302) -> -429 (5451:13753) 1169 hits
LongReadMapping 1000 (8530:14690) -> -4990 (190:6340) 963 hits
LongReadMapping 1000 (15831:20460) -> 2904 (8:4637) 611 hits
LongReadMapping 1000 (24032:25491) -> -4169 (14:1473) 213 hits
LongReadMapping 1000 (24032:25491) -> -2393 (14:1473) 259 hits
LongReadMapping 1000 (27748:29666) -> 2983 (21:1939) 240 hits
LongReadMapping 1000 (29531:30795) -> 4596 (12:1276) 215 hits
LongReadMapping 1000 (29531:30795) -> 4773 (12:1276) 127 hits
The same mappings can be applied to the rest of the datastores
## Map the rest of the ds in the workspace
>>> ws.get_paired_reads_datastore("PE").mapper.map_reads()
>>> ws.get_linked_reads_datastore("LI").mapper.map_reads()
Now that all the data is attached to the workspace and the reads are mapped, it is a good time to save the workspace.
ws.dump_to_disk("./persisted.reads.mapped.sdgws")
Now all the previous work is stored on disk and can be loaded when needed.
The ls method of a workspace is useful to show what is contained within the workspace;
print(ws.ls())
NodeViews
NodeViews are used to access the workspace data from a node perspective. For example, to create a nodeview of the 10th node of the main SDG graph in the workspace;
>>> nv = ws.sdg.get_nodeview(10)
Using nodeviews you can access node properties, navigate the graph, access datastores and mappings. For example, to see the properties of node 10;
## Get the node id
>>> print(nv.node_id())
10
## get the node sequence
>>> print(nv.sequence())
'AAAAAAAAAACTGTTAATTTTTTTTTTTACTGATTTACAAATTCCTTGTACCTCTATCTCAGCCCATGCATATATTGGTTGTTTTACTGCTTGATACCACTAGAAACCTCTAGTTGTTTAA'
## get the node size
>>> print(nv.size())
121
You can also query the topology of the graph from the 10th node perspective. For example, if you want to get the node neighbours;
>>> for n in nv.next():
>>> ....print(n)
<< NodeDistanceView: -60bp to 2 >>
<< NodeDistanceView: -60bp to -3 >>
>>> for p in nv.prev():
>>> ....print(p)
<< NodeDistanceView: ...bp to ... >>
<< NodeDistanceView: ...bp to ... >>
Mappings can also be queried from the nodeview,
## Get the names of the available linked reads datastore
>>> ws.list_linked_reads_datastores()
('LI',)
## get the id of the first 3 reads mapped to the node
>>> for m in nv.get_linked_mappings('LI')[:3]:
>>> ....print(m)
12576
23882
30795
## get the first 3 mappings from the datastore
>>> for m in nv.get_linked_mappings('LI')[:3]:
>>> ....print(m)
<pysdg.pysdg.ReadMapping; proxy of <Swig Object of type 'ReadMapping *' at 0x107bb5f90> >
<pysdg.pysdg.ReadMapping; proxy of <Swig Object of type 'ReadMapping *' at 0x107bb55d0> >
<pysdg.pysdg.ReadMapping; proxy of <Swig Object of type 'ReadMapping *' at 0x107bb5f90> >
Mappings contain the information to go from nodes to reads and vice versa, the information inside the mappings can be queried individually.
### Get the first mappings
>>> mapping = nv.get_linked_mappings('LI')[0]
## Check the node of the mapping
>>> mapping.node
10
## Check the read id
>>> mapping.read_id
12576
## Check the first and last mapping position in the node for the mapping
>>> mapping.first_pos
641
>>> mapping.last_pos
638
## Check the orientation of the mapping (True=opposite to node, False=Same as node)
>>> mapping.rev
Out[27]: True
Each data type has a slightly different mapping format.
Short reads (kci)
SDG can be also used to do kmer counting operations on the graph, SDG indexes the graph and then counts the kmers in any sequence collection. The data is stored in KmerCounter objects and similarly to the datastores can be attached to the workspace.
WARNING: To count faster and produce smaller hash files, SDG indexes the graph kmers in an internal index and will only consider kmers that are present in the original graph. This is something that should be kept in mind when doing kmer count analysis. If a motif is not represented in the main graph will not be present in any of the counts.
If you want to check the completeness of your graph you can use the KAT tool.
To count kmers you first need to add a KmerCounter object with some parameters such as the name of the collection, the k size and if you want to count canonical or non cannonical kmers and then add a count object inside the collection. For an explanation of canonical kmers go to this post.
>>> ws.add_kmer_counter("PEkmers", 21)
<KmerCounter PEkmers: index with 2266158 21-mers>
>>> ws.list_kmer_counters()
('PEkmers',)
## Count kmers directly from the pe .fastq files
>>> ws.get_kmer_counter("PEkmers").add_count('pe1', ['./pe-reads_R1.fastq', './pe-reads_R2.fastq'])
Now the counts are stored in the KmerCounter and can be accessed either via collection name or index.
>>> ws.get_kmer_counter('PEkmers') == ws.kmer_counters[0]
True
The counts inside the collection can also be queried using the KmerCounter object.
## List all kmer collections
>>> ws.get_kmer_counter('PEkmers').list_names()
('sdg', 'pe1')
## Find count in the library, the assembly
>>> kmers = ws.get_kmer_counter("pe1")
>>> kmers.project_count("pe1", "AAAAAAAAAAAAAAAAAAAAA")
(6332,)
If you want to check the kmer coverage for a node sequence you can use the sequence from the nodeview;
>>> kmers.project_count("pe1", ws.sdg.get_nodeview(10).sequence())
(126, 124, 126, 126, 126, 130, 130, 128, 128, 128, 128, 132, 132, 132, 130, 130, 132, 132, 132, 128, 126, 126, 126, 126, 122, 126, 126, 124, 122, 118, 118, 118, 118, 118, 118, 120, 120, 120, 120, 120, 120, 120, 122, 124, 128, 130, 58, 56, 56, 56, 56, 56, 56, 56, 54, 54, 54, 54, 54, 54, 54, 126, 124, 124, 124, 124, 126, 126, 128, 128, 128, 128, 128, 128, 128, 126, 126, 128, 128, 128, 128, 130, 130, 130, 132, 132, 130, 130, 186, 184, 134, 136, 136, 136, 134, 132, 132, 132, 134, 134, 134, 132, 134, 134, 134, 134, 134)
Or by directly accessing the count from the nodeview;
>>> nv = ws.sdg.get_nodeview(10)
>>> print(nv.kmer_coverage('kmers', 'pe1'))
(126, 124, 126, 126, 126, 130, 130, 128, 128, 128, 128, 132, 132, 132, 130, 130, 132, 132, 132, 128, 126, 126, 126, 126, 122, 126, 126, 124, 122, 118, 118, 118, 118, 118, 118, 120, 120, 120, 120, 120, 120, 120, 122, 124, 128, 130, 58, 56, 56, 56, 56, 56, 56, 56, 54, 54, 54, 54, 54, 54, 54, 126, 124, 124, 124, 124, 126, 126, 128, 128, 128, 128, 128, 128, 128, 126, 126, 128, 128, 128, 128, 130, 130, 130, 132, 132, 130, 130, 186, 184, 134, 136, 136, 136, 134, 132, 132, 132, 134, 134, 134, 132, 134, 134, 134, 134, 134)
Both functions return the kmer coverage for the given node.
For each kmer collection there is also kmer count that represents the count of the kmers in the main graph.